Foundation models for generalist medical artificial intelligence
Abstract
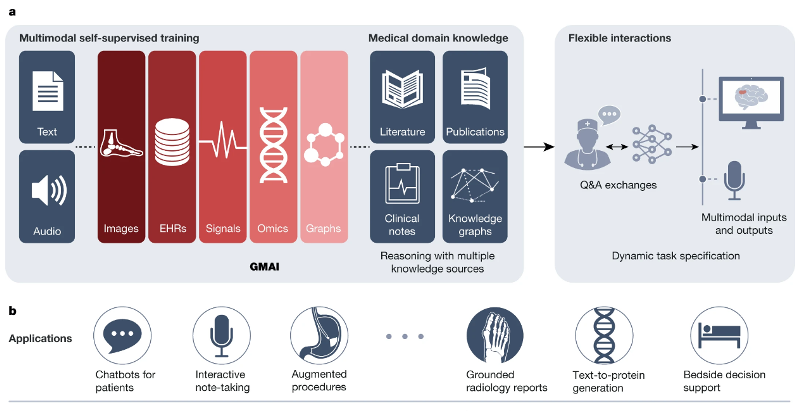
The exceptionally rapid development of highly flexible, reusable artificial intelligence (AI) models is likely to usher in newfound capabilities in medicine. We propose a new paradigm for medical AI, which we refer to as generalist medical AI (GMAI). GMAI models will be capable of carrying out a diverse set of tasks using very little or no task-specific labelled data. Built through self-supervision on large, diverse datasets, GMAI will flexibly interpret different combinations of medical modalities, including data from imaging, electronic health records, laboratory results, genomics, graphs or medical text. Models will in turn produce expressive outputs such as free-text explanations, spoken recommendations or image annotations that demonstrate advanced medical reasoning abilities. Here we identify a set of high-impact potential applications for GMAI and lay out specific technical capabilities and training datasets necessary to enable them. We expect that GMAI-enabled applications will challenge current strategies for regulating and validating AI devices for medicine and will shift practices associated with the collection of large medical datasets.
Main
Foundation models—the latest generation of AI models—are trained on massive, diverse datasets and can be applied to numerous downstream tasks. Individual models can now achieve state-of-the-art performance on a wide variety of problems, ranging from answering questions about texts to describing images and playing video games. This versatility represents a stark change from the previous generation of AI models, which were designed to solve specific tasks, one at a time.
Driven by growing datasets, increases in model size and advances in model architectures, foundation models offer previously unseen abilities. For example, in 2020 the language model GPT-3 unlocked a new capability: in-context learning, through which the model carried out entirely new tasks that it had never explicitly been trained for, simply by learning from text explanations (or ‘prompts’) containing a few examples. Additionally, many recent foundation models are able to take in and output combinations of different data modalities. For example, the recent Gato model can chat, caption images, play video games and control a robot arm and has thus been described as a generalist agent. As certain capabilities emerge only in the largest models, it remains challenging to predict what even larger models will be able to accomplish.
Although there have been early efforts to develop medical foundation models, this shift has not yet widely permeated medical AI, owing to the difficulty of accessing large, diverse medical datasets, the complexity of the medical domain and the recency of this development. Instead, medical AI models are largely still developed with a task-specific approach to model development. For instance, a chest X-ray interpretation model may be trained on a dataset in which every image has been explicitly labelled as positive or negative for pneumonia, probably requiring substantial annotation effort. This model would only detect pneumonia and would not be able to carry out the complete diagnostic exercise of writing a comprehensive radiology report. This narrow, task-specific approach produces inflexible models, limited to carrying out tasks predefined by the training dataset and its labels. In current practice, such models typically cannot adapt to other tasks (or even to different data distributions for the same task) without being retrained on another dataset. Of the more than 500 AI models for clinical medicine that have received approval by the Food and Drug Administration, most have been approved for only 1 or 2 narrow tasks.
Here we outline how recent advances in foundation model research can disrupt this task-specific paradigm. These include the rise of multimodal architectures and self-supervised learning techniques that dispense with explicit labels (for example, language modelling and contrastive learning), as well as the advent of in-context learning capabilities.
These advances will instead enable the development of GMAI, a class of advanced medical foundation models. ‘Generalist’ implies that they will be widely used across medical applications, largely replacing task-specific models.
Inspired directly by foundation models outside medicine, we identify three key capabilities that distinguish GMAI models from conventional medical AI models. First, adapting a GMAI model to a new task will be as easy as describing the task in plain English (or another language). Models will be able to solve previously unseen problems simply by having new tasks explained to them (dynamic task specification), without needing to be retrained. Second, GMAI models can accept inputs and produce outputs using varying combinations of data modalities (for example, can take in images, text, laboratory results or any combination thereof). This flexible interactivity contrasts with the constraints of more rigid multimodal models, which always use predefined sets of modalities as input and output (for example, must always take in images, text and laboratory results together). Third, GMAI models will formally represent medical knowledge, allowing them to reason through previously unseen tasks and use medically accurate language to explain their outputs.
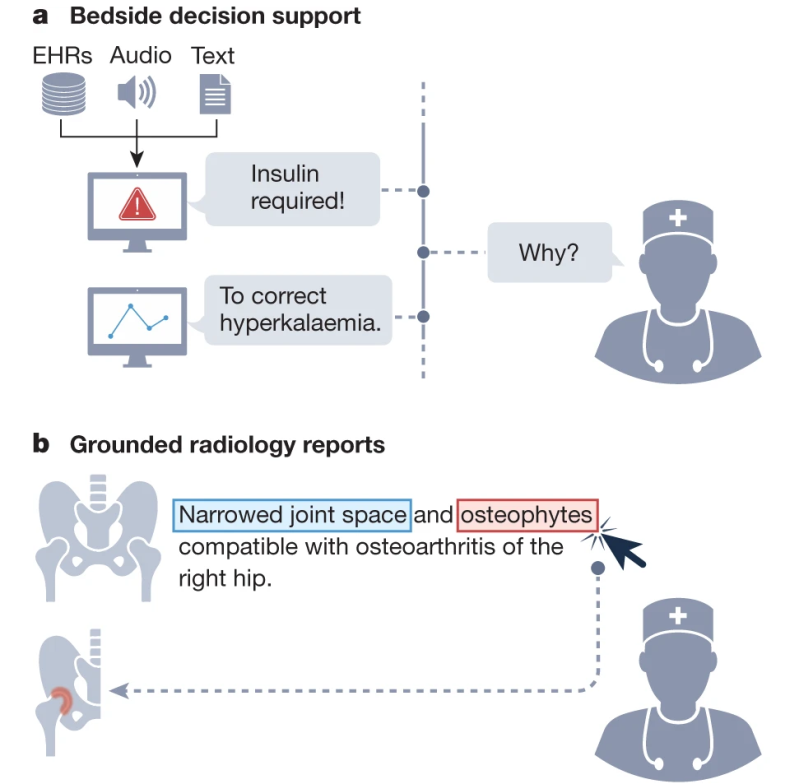
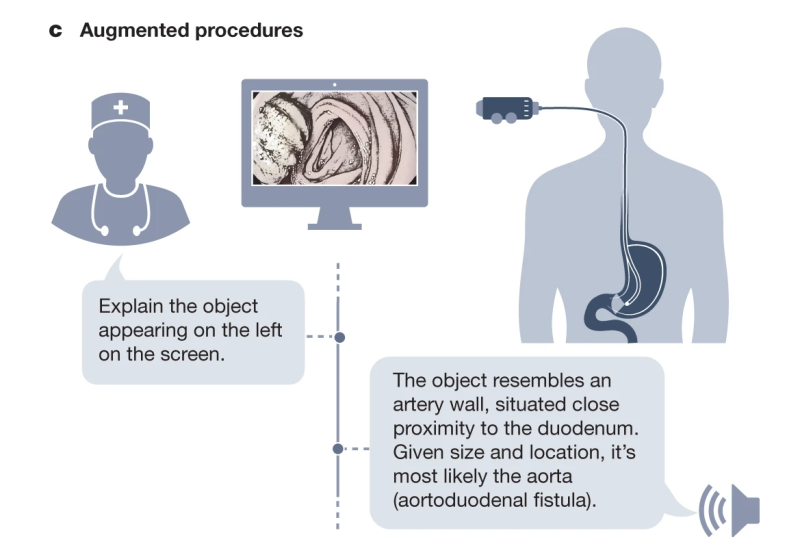
We list concrete strategies for achieving this paradigm shift in medical AI. Furthermore, we describe a set of potentially high-impact applications that this new generation of models will enable. Finally, we point out core challenges that must be overcome for GMAI to deliver the clinical value it promises.
For More information, please visit: https://www.nature.com/articles/s41586-023-05881-4
Sources: Nature volume 616, pages259–265 (2023)
Published: 12 April 2023
DOI: 10.1038/s41586-023-05881-4
Disclaimer:
Partial content of this page is transferred from the network, only for the use of scientific communication, if there is infringement, please contact us to delete. See the Privacy Policy for more information.